MBBC V1.1 Manual
1. Prerequisites
MBBC can be run on systems with Java installed:** Java 7 Runtime Environment.
You need to install Java 7 (64-bit is highly recommended). To check the version of the Java installed, please input the following command in the terminal:
java -versionThe latest version can be downloaded here.
Please make sure that java is available and can be accessed from the command prompt if you run MBBC on Windows, otherwise please add java path to the Environment variables. Be default, java will be installed in "c:\Program Files\Java\jre7\bin\java", so you can run java by using command as following: For 32-bit java:
"c:\Program Files (x86)\Java\jre7\bin\java" -versionOr for 64-bit java:
"c:\Program Files\Java\jre7\bin\java" -version** We use KAnalyze to do the k-mer counting.
This tool was included in the software package.
Audano, Peter, and Fredrik Vannberg. "KAnalyze: a fast versatile pipelined K-mer toolkit." Bioinformatics 30.14 (2014): 2070-2072.
2. Input Parameters
There are only three parameters for MBBC.-
Reads files: paired-end or single-end reads in FASTA format
Paired-end reads should be in following format:
>Read1_end1
ACGT...
>Read1_end2
ACGT...
>Read2_end1
ACGT...
>Read2_end2
ACGT...
- initial species number m (should be larger than the real species number), by default m=10.
-
Read type: 0 or 1, by default Read type=0;
0: denotes paired-end reads;
1: denotes single-end reads;
3. Software Usage
We have both CLI (command line) and GUI versions for MBBC on Linux and Window.(1) Command line versions (CLI) of MBBC
In terminal, use the following to run MBBC:
java -jar -XmxN MBBC.jar -i reads_files -m species_number -r read_type(0 or 1)
- -XmxN(for example:-Xmx7g): Specify the maximum size of the memory allocation pool for a Java Virtual Machine (JVM)
- -i: reads files in FASTA format (Required)
- -m: species number, by default m=10 (Optional)
- -r: 0: paired_end reads; 1: single-end reads, be default r=0 (Optional)
(2) GUI versions of MBBC
Run the following command in the terminal:
java -jar -XmxN MBBC_GUI.jar
-XmxN(for example:-Xmx7g): Specify the maximum size of the memory allocation pool for a Java Virtual Machine (JVM)
This will open a window where you can input the above three parameters, then you just need to click 'Run'. When MBBC is running, the running progress is shown in the 'Outputs' window; after MBBC is done, it will generated output files.
Note: You can also double-click the MBBC executable to run it, but with this you can't specify enough memory so you might run into the memory allocation error like this:"Exception in thread "Thread-0" java.lang.OutOfMemoryError: GC overhead limit exceeded".
The following graph shows an example of GUI verions of MBBC:
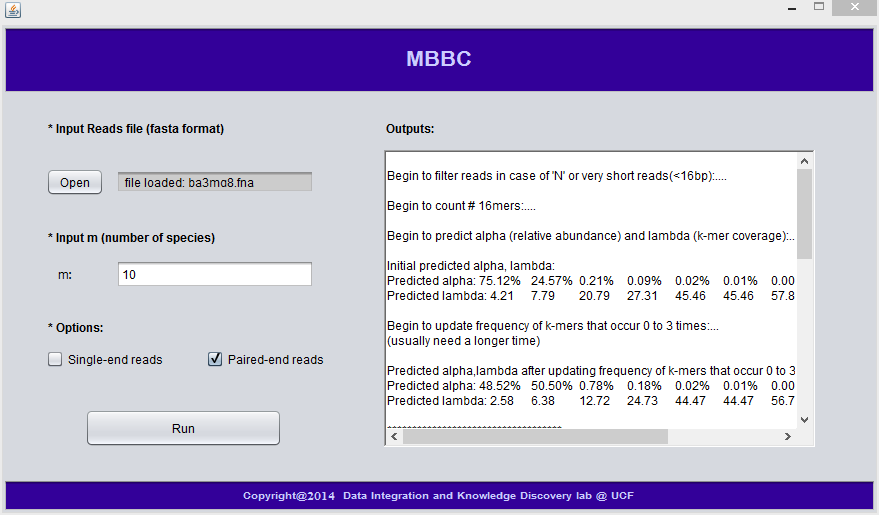
4. MBBC Results
We take dataset "ba3mg8.fna" as an example, which contains two species, and genome coverage ratio is 3:8.We can run MBBC as the following command:
java -jar -Xmx7g MBBC.jar -i MBBC_examples/paired-end_reads/ba3mg8.fna
(1) Output process:
This will give you a general look of the process when MBBC runs.
Example output process is as following:
Begin to filter reads in case of 'N' or very short reads(<16bp):.... Begin to count # 16mers:.... Begin to predict alpha (relative abundance) and lambda (k-mer coverage):.... Initial predicted alpha, lambda: Predicted alpha: 75.20% 24.51% 0.19% 0.07% 0.02% 0.01% 0.00% 0.00% 0.00% 0.00% Predicted lambda: 4.20 7.76 21.53 27.59 45.33 45.33 57.58 57.58 57.58 186.00 Begin to update frequency of k-mers that occur 0 to 3 times:... (usually need a longer time) Predicted alpha,lambda after updating of k-mers that occur 0 to 3 times:... Predicted alpha: 48.83% 50.33% 0.65% 0.17% 0.02% 0.00% 0.00% 0.00% 0.00% 0.00% Predicted lambda: 2.59 6.38 12.79 24.83 44.39 44.39 56.52 56.52 56.52 186.00 *********************************** Predicted genome size: 580108.91 577958.56 Predicted alpha: 28.93% 71.07% Predicted lambda: 2.59 6.38 There are 2 species predicted. *********************************** Assign reads to m groups.... Start training Markov Chain until it coverage.... loop 0.... Write reads into 2 files.... *********************************** Begin to predict the genome size, alpha from reads assignments... Finally predicted genome size: 522830.37 601189.22 Predicted alpha: 26.07% 73.93% Predicted lambda: 2.59 6.38 *********************************** The program is finished.. Total running time: 1.57 mins(2) Output files:
Example output files are as following:
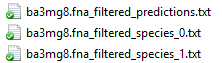
The first file contains the predicted species number, genome sizes, k-mer converge and relative abundance.
Example file is as following, which showed the predicted parameters before and after reads assignments.
Predictions after EM algorithms: Predicted genome size: 580108.91 577958.56 Predicted alpha: 28.93% 71.07% Predicted lambda: 2.59 6.38 There are 2 species predicted. Prediction after reads binning: Predicted genome size: 522830.37 601189.22 Predicted alpha: 26.07% 73.93% Predicted lambda: 2.59 6.38
Other output files were binned reads in each of separate files in FASTA format.